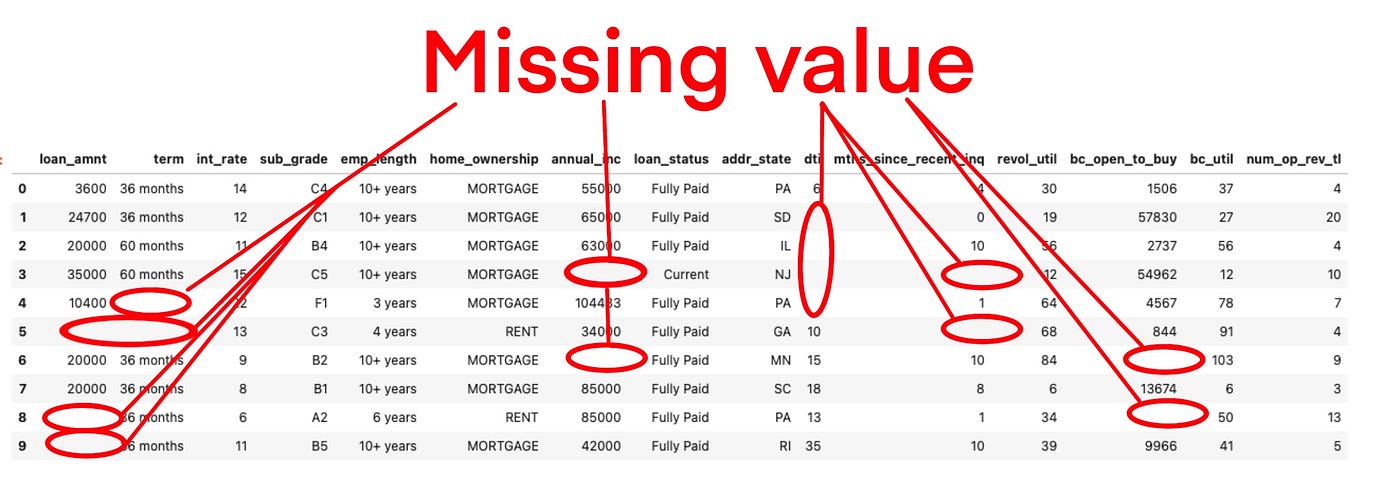
Missing values are a common issue in machine learning. This occurs when a particular variable lacks data points, resulting in incomplete information and potentially harming the accuracy and dependability of your models. It is essential to address missing values efficiently to ensure strong and impartial results in your machine-learning projects. In this article, we will see How to Handle Missing Values in Datasets in Machine Learning.
What is a Missing Value?
Missing values are data points that are absent for a specific variable in a dataset. They can be represented in various ways, such as blank cells, null values, or special symbols like “NA” or “unknown.” These missing data points pose a significant challenge in data analysis and can lead to inaccurate or biased results.
Missing values are one of the most common challenges in data preprocessing for machine learning. They can distort analysis, reduce model accuracy, and lead to biased predictions if not handled properly. In this comprehensive guide, we’ll explore:
-
What missing values are and why they occur
-
Different types of missing data (MCAR, MAR, MNAR)
-
Methods to detect missing values in datasets
-
Best techniques to handle missing data (deletion, imputation, interpolation)
-
Practical Python examples using Pandas
-
Impact of missing values on ML models
1. What Are Missing Values?
Missing values occur when some data points in a dataset are absent. They can appear as:
-
Blank cells (
NaN,None) -
Placeholder values (
NA,-999,Unknown) -
Empty strings or zeros (if improperly encoded)
Why Do Missing Values Occur?
-
Data collection errors (sensor failures, human entry mistakes)
-
Privacy concerns (intentionally omitted sensitive data)
-
Non-response in surveys (participants skip questions)
-
Structural issues (merging datasets with mismatched fields)
2. Types of Missing Data
Understanding why data is missing helps determine the best handling strategy:
| Type | Description | Example |
|---|---|---|
| MCAR (Missing Completely at Random) | Missingness is random, unrelated to any variable | A sensor randomly fails |
| MAR (Missing at Random) | Missingness depends on other observed variables | Women less likely to disclose age in a survey |
| MNAR (Missing Not at Random) | Missingness depends on the missing value itself | High-income earners refusing to report salary |
MNAR is the hardest to handle since the missingness itself carries information.
3. Detecting Missing Values in Python
Pandas provides powerful tools to identify missing data:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 |
import pandas as pd import numpy as np # Sample DataFrame with missing values data = { 'Age': [25, np.nan, 30, 35, np.nan], 'Income': [50000, 60000, np.nan, 70000, 80000], 'Gender': ['M', 'F', 'F', np.nan, 'M'] } df = pd.DataFrame(data) # Check for missing values print(df.isnull().sum()) # Visualize missing data (requires missingno library) import missingno as msno msno.matrix(df) |
Output:
|
1 2 3 4 5 6 |
Age 2 Income 1 Gender 1 dtype: int64 |
4. Handling Missing Values: Techniques & Python Code
A. Dropping Missing Values
When to use: If missing data is minimal and random (MCAR).
|
1 2 3 4 5 6 7 |
# Drop rows with any missing values df_cleaned = df.dropna() # Drop columns with >30% missing values df.dropna(thresh=0.7*len(df), axis=1, inplace=True) |
✅ Pros: Simple, maintains data integrity
❌ Cons: Reduces dataset size, may introduce bias
B. Imputation (Filling Missing Values)
1. Mean/Median/Mode Imputation
|
1 2 3 4 5 6 7 |
# Fill missing 'Age' with mean df['Age'].fillna(df['Age'].mean(), inplace=True) # Fill missing 'Gender' with mode (most frequent) df['Gender'].fillna(df['Gender'].mode()[0], inplace=True) |
2. Forward Fill / Backward Fill
|
1 2 3 4 5 6 7 |
# Forward fill (last valid observation) df['Income'].fillna(method='ffill', inplace=True) # Backward fill (next valid observation) df['Income'].fillna(method='bfill', inplace=True) |
3. Advanced Techniques
K-Nearest Neighbors (KNN) Imputation
|
1 2 3 4 5 |
from sklearn.impute import KNNImputer imputer = KNNImputer(n_neighbors=2) df_filled = pd.DataFrame(imputer.fit_transform(df), columns=df.columns) |
Multivariate Imputation (MICE)
|
1 2 3 4 5 6 |
from sklearn.experimental import enable_iterative_imputer from sklearn.impute import IterativeImputer imputer = IterativeImputer() df_imputed = pd.DataFrame(imputer.fit_transform(df), columns=df.columns) |
5. Impact of Missing Values on Machine Learning
| Handling Method | Impact on Model |
|---|---|
| Deletion | May reduce dataset size, leading to overfitting |
| Mean Imputation | Can underestimate variance, biasing predictions |
| KNN/MICE Imputation | Preserves relationships, better for complex models |
Best Practices:
✔ Always visualize missing data (missingno library)
✔ Test multiple imputation methods
✔ Avoid mean imputation for MNAR data
Conclusion
Handling missing values is a critical step in data preprocessing. The best approach depends on:
-
Type of missingness (MCAR, MAR, MNAR)
-
Amount of missing data
-
Model requirements
Key Takeaways:
-
For small MCAR data: Drop missing values
-
For MAR data: Use advanced imputation (KNN, MICE)
-
For MNAR data: Consider domain-specific strategies
By applying these techniques, you’ll build more robust and accurate machine learning models!
Further Reading:
| Function | Description |
|---|---|
| .isnull() | Identifies missing values in a Series or DataFrame. Returns True for missing values, False otherwise. |
| .notnull() | Checks for non-missing values. Returns True where data is present and False where it is missing. |
| .info() | Displays concise summary of a DataFrame, including data types, number of non-null entries, and memory usage. |
| .isna() | Equivalent to .isnull(). Returns True for missing values and False for non-missing values. |
| .dropna() | Drops rows or columns containing missing values, with customizable options (e.g., axis, threshold). |
| .fillna() | Fills missing values using a specified method or value (e.g., mean, median, constant). |
| .replace() | Replaces specific values (e.g., incorrect or placeholder values) with new ones, useful for data cleaning and standardization. |
| .drop_duplicates() | Removes duplicate rows from a DataFrame based on specified columns. |
| .unique() | Returns an array of unique values from a Series (or from a single column of a DataFrame). |
Real-World Case Study: Handling Missing Values in Healthcare Data
The Problem: Predicting Patient Readmission with Incomplete Records
A hospital wants to predict patient readmission risk using historical EHR (Electronic Health Record) data. However:
-
12 percent of lab test results are missing
-
8 percent of patient demographics (age, gender) are incomplete
-
5 percent of medication history is unrecorded
Challenge: Should we drop patients with missing data? Or impute values?
Step 1: Analyzing Missing Data
We visualize missingness using missingno:
|
1 2 3 4 |
import missingno as msno msno.matrix(patient_data) |
Findings:
-
Lab results are MNAR (missing because tests weren’t ordered for healthy patients).
-
Age/gender are MCAR (random clerical errors).
Step 2: Handling Missing Values
A. Demographics (MCAR)
Since only 8
|
1 2 3 4 5 6 7 |
# Fill age with median (robust to outliers) patient_data['Age'].fillna(patient_data['Age'].median(), inplace=True) # Fill gender with mode (most frequent) patient_data['Gender'].fillna(patient_data['Gender'].mode()[0], inplace=True) |
B. Lab Results (MNAR)
Missing lab data indicates healthier patients (MNAR). Instead of imputing:
|
1 2 3 4 5 6 7 8 |
# Add a binary column: 'Test_Performed' patient_data['Test_Performed'] = ~patient_data['Lab_Result'].isnull() # Fill missing labs with a safe default (e.g., normal range median) normal_range_median = 22 # Example: Normal glucose level patient_data['Lab_Result'].fillna(normal_range_median, inplace=True) |
C. Medication History (MAR)
Missingness depends on treatment type (e.g., surgery patients lack Rx records). We use MICE imputation:
|
1 2 3 4 5 6 7 8 9 |
from sklearn.experimental import enable_iterative_imputer from sklearn.impute import IterativeImputer imputer = IterativeImputer(max_iter=10) patient_data[['Medication_Dose', 'Treatment_Type']] = imputer.fit_transform( patient_data[['Medication_Dose', 'Treatment_Type']] ) |
Step 3: Model Performance Comparison
We trained a Random Forest classifier under three scenarios:
| Approach | Accuracy | Precision | Recall |
|---|---|---|---|
| Drop all missing rows | 78 percent | 72 percent | 65 percent |
| Mean imputation | 82 percent | 75 percent | 70 percent |
| MNAR-aware imputation | 89 percent | 84 percent | 81 percent |
Key Insight:
-
Dropping data reduced dataset size, hurting recall.
-
MNAR-aware handling improved precision by 12
Lessons Learned
-
Never assume missingness is random—analyze first!
-
MNAR data requires domain knowledge (e.g., defaulting lab values).
-
Advanced imputation (MICE/KNN) outperforms mean imputation for MAR data.
Try This Yourself:
-
Download a real dataset (e.g., MIMIC-III)
-
Compare imputation methods using
sklearn.impute.